BIO392 - Bioinformatics of Sequence Variation
One of the fastest growing areas of bioinformatics is in the analysis, warehousing and representation of genomic and protein sequence variants, particularly with view on the use of molecular data in personalised health and biomedical applications in general. This course will engage participants to explore common data formats, online resources and analysis techniques, with a focus on human genome variation data.
Practical Information
- from 2025 on in each Spring semester (4 weeks in May) instead of Autumn semester
- Vorlesungsverzeichnis (FS26)
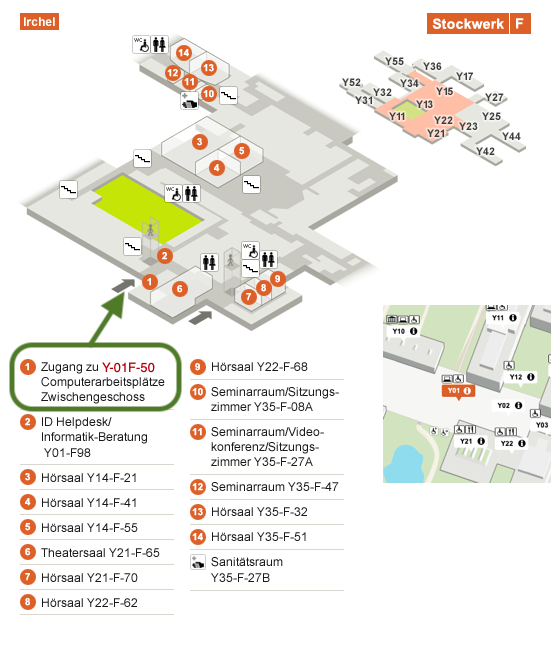
- UZH Irchel campus, Y-01F-50
- Course language is English
- OLAT
- previous BIO392 HS 2023 in the UZH OLAT system
- compbiozurich/UZH-BIO392 course material Github repository
- you will need to have / create a Github account (though this is also addressed at the beginning of the course)
- (might change for 2026) the course lab is equipped with iMacs - the use your personal machines is discouraged unless you really know what to do...
Location
Course Days
Exam & Course Review
UZH BIO392 FS25 - Day 13
Supervision: Michael
Time: 09:00 - 12:00
On the last day of the course there will be a 1:30h written exam which will consist of multiple choice questions and 2-3 open essay questions.
For preparation, you should be aware of are mostly the basics of the individual topics of the course, e.g. (non-exclusive examples) Continue reading
Genomic Data - Genomic Privacy
UZH BIO392 FS25 - Day 12
Supervision: Michael Baudis
Time: 13:30 - 16:30
The last day of the course presents & discsusses topics somehow related to genomic privacy, including the following:
Continue readingTBD
UZH BIO392 FS25 - Day 11
Supervision: TBD
Time: 09:00 - 16:30
TBD (home study)
Continue readingProjects
UZH BIO392 FS25 - Day 07
Supervision: Feifei Xia
Time: 09:00 - 16:30
Work on the project as a group of two
docs.google.com/spreadsheets/d/1--S8DInxkt8H1jW7CIt2SWFTpnmNp3jGszimGg7jkc4/edit?usp=sharing
Continue readingIntroduction to STRs
UZH BIO392 FS25 - Day 07
Supervision: Feifei Xia
Time: 13:00 - 16:30
- STR genotyping exercise
Data Sharing, Beacon, ...
UZH BIO392 FS25 - Day 08
Supervision: Miochael Baudis
Time: 09:00 - 12:00 (afternoon for reading etc.)
- some survival analysis theory
- cancer classifications
- data sharing, Progenetix, EGA...
- GA4GH and Beacon
Introduction to STRs
UZH BIO392 FS25 - Day 07
Supervision: Feifei Xia
Time: 09:00 - 16:00
- Introduction to STRs
- STR genotyping exercise
Introduction to SNPs
UZH BIO392 FS25 - Day 06
Supervision: Feifei Xia
Time: 13:00 - 16:30
Introduction to SNPs
Introduction to the GWAS catalog
Genomic Resources, Data Sharing, and Introducing BLAST Basics
UZH BIO392 FS25 - Day 05
Supervision: Michael Baudis, Hangjia Zhao
Time: 09:00 - 16:30
- some genomic file formats
- online resources for genome data
- hands-on exercises using these resources and an introduction to BLAST
Basics of R Programming with a Case Study in CNV Analysis
UZH BIO392 FS25 - Day 04
Supervision: Hangjia Zhao
Time: 09:00 - 16:30
Learn the basics of R programming and apply them in a hands-on case study focused on copy number variation (CNV) analysis
Continue readingUnix & Python Introduction
UZH BIO392 FS25 - Day 03
Supervision: Jiahui, Ziying
Time: 09:00 - 16:30
Learn the basics of Unix (systems, filesystem, process commands, and basic file management, including setting file permissions). Learn the basics of Python programming and apply them in a hands-on case study focused on copy number variation (CNV) analysis.
Continue readingAnalyses of Human Genome Variation
An Introduction
UZH BIO392 FS25 - Day 02
Supervision: Michael Baudis
Time: 13:00 - 16:30
The understanding of the human genome with respect to both inherited (germline) and acquired (somatic) variation is a key to understanding human biology and diseases.
Continue readingIntroduction & Technicalities
UZH BIO392 FS25 - Day 01
Supervision: TBD
Time: 09:00 - 16:30
The first day of the "Bioinformatics of Sequence Variation" course starts with a general introduction to the course's topics, timeline and procedures.
Continue readingGenomic Data & Privacy - Risks and Opportunities
UZH BIO392 HS22 - Day 12
Michael Baudis
Today we will discuss opportunities and Risks from genomic data collections and general privacy implications of genomic and other molecular screening data.
Continue readingSequence analysis: interpretation
UZH BIO392 HS23 - Day 10
Morning
On the morning of day 10, the output of day 08's practical session (i.e., variant calls) will be analysed interpreted. Which variants were observed in the sequencing data you were provided and how can this be related to existing knowledge?
Continue readingSurvival Analysis Exercise & Discussion
UZH BIO392 HS22 - Day 12
Survival analysis
This day and the following day of the course we will be working with survival datasets. Plan:
- Introduction
- Form groups (2-3 people)
- Data download and prep
- Work in groups
Survival as a Measure in Cancer Genomics? Also: Cancer Classifications
UZH BIO392 HS22 - Day 09
Michael Baudis
This day provides some notes about the submitted notes, introduction to Kaplan-Meier survival analysis concepts and classification systems used to describe cancer types and clinical apperances.
Continue readingSequence analysis: practical
UZH BIO392 HS23 - Day 08
Morning
The morning of day 08 will feature a hands-on practical of the bioinformatics pipeline discussed on the afternoon of day 07. The day will start with a brief refresher on the starting point of our pipeline: files with raw sequencing reads and a reference genome. After this, time will be spent on generating alignments and variant calls from the provided data, applying several widely used bioinformatics tools along the way.
Afternoon
The afternoon will be spent on reflecting on (or finishing) the steps from the practical. Afterwards, literature and/or slides will be provided to provide more background on Short Tandem Repeats (STRs); What are they? Why can they be problematic to analyse? Why is their variation relevant? These questions should prepare you for the morning of day 10, where the outputs of the bioinformatics pipeline will be analyses and interpreted.
Continue readingGenomic resource usage, sequence comparison, and sequence analysis
UZH BIO392 HS23 - Day 07
On this day, we will do hands-on exercises using some clinical variant annotation resources, and provide an overview of BLAST and a typical bioinformatics pipeline.
Continue readingProgenetix, Beacon+, EGA and Genome Resources
UZH BIO392 HS23 - Day 06
Michael Baudis
Today will be presentations about several topics, including Data Federation through the Beacon protocol as well as the Progenetix database. Another presentation will point to some online resources for genomics data.
Continue readingGenome Analysis Technologies
UZH BIO392 HS23 - Day 05
Michael Baudis
Some information about genomic analysis technologies, file formats ...
Continue readingGenomic File Formats, 1000 genomes project
UZH BIO392 HS22 - Day 04
Izaskun Mallona (email: izaskun.mallona at sib.swiss).
Morning
We will have a lecture and run a set of exercises (on site).
- Overview of the standard genomics data formats
- FASTA
- FASTQ
- SAM
- BED
- GFF
- VCF
- Basic file processing for bioinformatics
- wc, grep, awk
- Exercises
- Project
Afternoon
Exercises and project.
- SAM v1 format specification
- BEDtools paper
- 0-start, 1-start, open, closed: how do we count
- GFF3 format
- VCF format
Terminal, Unix & Files
UZH BIO392 HS23 - Day 03
Izaskun Mallona (email: izaskun.mallona at mls.uzh.ch).
Morning (on site)
We will have two lectures and several sets of exercises (on site).
- Unix + genomic formats lectures (exercises 1-4)
- Unix and genomic formats exercises (5-14)
Afternoon
We will run the SIB Unix course using a Web browser. We encourage you to run the exercises of the course in a terminal of your own; either on a GNU/Linux or MacOS; or with a Web browser-based emulator of your choice, like cocalc.
- SIB Unix course: UNIX for Bioinformatics
- Chapter 1: What is UNIX
- Chapter 2: The UNIX filesystem
- Chapter 3: UNIX shell - first steps
- Chapter 4: UNIX shell - filesystem commands
- Chapter 5: UNIX shell - working with files
Tools for programming
UZH BIO392 HS23 - Day 02
This day will introduce some tools for better software development such as Github and Atom, and basics of Python & R.
Continue readingIntroduction & Technicalities
UZH BIO392 HS23 - Day 01
The first day of the "Bioinformatics of Sequence Variation" course (afternoon only) starts with a general introduction to the course's topics, timeline and procedures.
Continue readingTest...
UZH BIO392 HS22 - Day 14
Continue readingGenomic Privacy and Q & A
UZH BIO392 HS22 - Day 13
Michael Baudis
Continue readingSurvival Analysis Exercise & Discussion
UZH BIO392 HS22 - Day 12
Continue readingSurvival Analysis Exercise & Discussion
UZH BIO392 HS22 - Day 11
Continue readingFrom Sequence to Variants & Survival Analysis Introduction
UZH BIO392 HS22 - Day 10
Continue readingSurvival as a Measure in Cancer Genomics? Also: Cancer Classifications
UZH BIO392 HS22 - Day 09
Michael Baudis
This day provides some notes about the submitted notes, introduction to Kaplan-Meier survival analysis concepts and classification systems used to describe cancer types and clinical apperances. Continue reading
Sequences Repeating Themselves - STRs et al.
UZH BIO392 HS22 - Day 08
Continue readingSequence Search with BLAST & Clinical Variant Interpretation Resource
UZH BIO392 HS22 - Day 07
Morning
-
introduction of BLAST
-
excercise on using BLAST
Afternoon
get familiar with some clinical variant annotation resources.
-
introduction of ClinVar and ClinGen
-
exercise on using ClinVar and ClinGen
Online Resources for Human Genomic Variations & Introduction to CNVs
UZH BIO392 HS22 - Day 06
Continue readingGenome Analysis Technologies
UZH BIO392 HS22 - Day 05
Michael Baudis
Some information about genomic analysis technologies, file formats ...
Continue readingGenomic File Formats, 1000 genomes project
UZH BIO392 HS22 - Day 04
Izaskun Mallona (email: izaskun.mallona at sib.swiss).
Morning (on site, 9 am)
We will have a lecture and run a set of exercises (on site).
- Overview of the standard genomics data formats
- FASTA
- FASTQ
- SAM
- BED
- GFF
- VCF
- Basic file processing for bioinformatics
- wc, grep, awk
- Exercises
- Project
Afternoon (on site or online)
Exercises and project.
- SAM v1 format specification
- BEDtools paper
- 0-start, 1-start, open, closed: how do we count
- GFF3 format
- VCF format
Terminal, Unix & Files
UZH BIO392 HS22 - Day 03
Izaskun Mallona (email: izaskun.mallona at mls.uzh.ch).
Morning (on site, 9 am)
We will have two lectures and several sets of exercises (on site).
- Unix + genomic formats lectures (exercises 1-4)
- Unix and genomic formats exercises (5-14)
Afternoon (on site or online)
We will run the SIB Unix course using a Web browser. We encourage you to run the exercises of the course in a terminal of your own; either on a GNU/Linux or MacOS; or with a Web browser-based emulator of your choice, like cocalc. Continue reading
Github, Terminal & Editors
UZH BIO392 HS22 - Day 02
The second day of the "Bioinformatics of Sequence Variation" course, with some inroduction and setups of Github, Terminal & Editors.
- 9:00 - 10:00: Introduction of Github things.
- 10:00 - 11:00: Github exercise: create user specific directories & upload/edit test files using Markdown.
-
11:00 - 12:00: Introduction to different interfaces eg atom, pycharm (lecture), editor setup exercise.
-
13:00 - 14:00: Get familiar with R (lecture).
- 14:00 - 15:00: Literature (genome variant review papers).
- 15:00 - 16:30: Task: Answer the questions of literature review session and provide some notes (1-2 pages total) in a doc posted on Github (.md)
Introduction & Technicalities
UZH BIO392 HS22 - Day 01
The first day of the "Bioinformatics of Sequence Variation" course (afternoon only) starts with a general introduction to the course's topics, timeline and procedures.
Continue reading